D-Aspartate oxidase
Degrading D-aspartate
In mammals, D-aspartate (D-Asp) shows a peculiar spatial distribution and temporal pattern of emergence [Errico et al., 2015a]. It is mainly present in the central nervous system, the neuroendocrine system, testis, and spermatozoa. Accordingly, this D-amino acid has been correlated to several crucial processes, such as development and neurogenesis of several areas of the brain (it is present at high concentrations in the central nervous system during the development of the fetus) and also in fertility.
The peculiar distribution of D-Asp requires an efficient and tightly controlled biochemical metabolism for its synthesis and degradation. The biochemical pathway for producing D-Asp in mammals is still under debate, but there is little doubt that the flavoprotein D-aspartate oxidase (DASPO or DDO, which was identified for the first time in 1949 in rabbit kidney) is the principal enzyme responsible for the catabolic metabolism of D-Asp [Katane et al., 2010a].
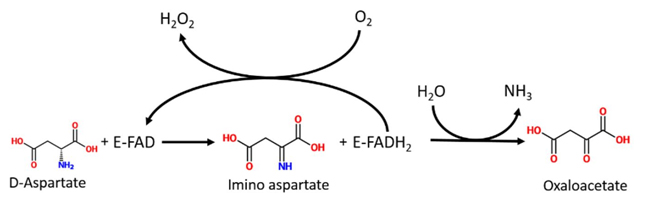
DASPO (EC 1.4.3.1) is a peroxisomal flavoenzyme which catalyzes the oxidative deamination of D-aspartate to imino aspartate which, in water, spontaneously deaminates, producing oxaloacetate and ammonia. The reduced cofactor is reoxidized by molecular oxygen with the production of a molecule of hydrogen peroxide (Fig. 1).
Distribution and physiological role
DASPO is present not only in mammals, but in almost all animals, from marine invertebrates to primates; accordingly, DASPO activity has been detected in several organisms, e.g., in cephalopods, gastropods, fishes, amphibians, birds, mammals, and yeast [Setoyama et al., 1997; Negri et al., 1999]. Interestingly, DASPO is not present in bacteria or plants, an aspect that connects this enzyme to D-amino acid oxidase, which shows a similar distribution in nature. Accordingly, phylogenetic analysis shows that genes coding for these two proteins are paralogues (i.e., they originated via a gene duplication event early in evolution).
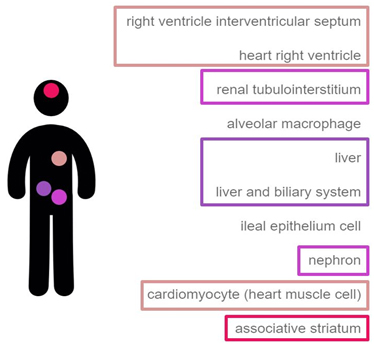
In mammals, DASPO shows a distribution inversely correlated to that of D-Asp; this enzyme is mainly present in heart muscle, kidney (epithelial cells of proximal renal tubules), liver (hepatocytes), and brain (Fig. 2) [https://genevisible.com/tissues/HS/Gene%20Symbol/DDO]. While in kidney the main physiological role of DASPO is probably to catabolically oxidize D-Asp deriving from spontaneous racemization or diet intake, in brain the enzyme is probably involved in the fine tuning of D-Asp (and possibly NMDA) local concentration [Katane et al., 2010; Molla, unpublished].
Biochemical properties
Most DASPOs are monomeric proteins; only few DASPOs have been reported as tetrameric proteins. The protein noncovalently binds a molecule of FAD with a Kd value ranging from ~ 10-6 M (mouse) to 3×10-8 M (human) and even ~ 10-10 M (yeast). The human enzyme is quite stable, having a Tm of 49.4 °C, and it is active in the pH range 8-12. Its stability is increased by the presence of ligands at the active site, a feature common to other amino acid oxidases.
The substrate specificity among mammalian DASPOs is similar: the preferred substrates are D-aspartate (Km = 1.1 mM, kcat 43.3 s-1, human enzyme) and NMDA. The enzyme is also active, even to a very low extent, on D-Glu and D-Asn, but with an efficiency that is less than 10% in comparison with D-Asp [Katane et al., 2015a; Molla, unpublished]. Importantly, DASPO is not active on the other two most important free amino acids in brain: glycine and D-serine. Nonmammalian DASPOs show a very different pattern of substrate specificity, probably related to different specific roles.
Mammalian DASPOs are very similar from a structural point of view, too (Fig. 3). They share a degree of sequence identity ranging from 75% to 91%. The experimental structure of mammalian DASPO is lacking; nonetheless, homology models show that the general setup of the active site of DASPO is very similar to that of DAAO [Umhau et al., 2000; Pollegioni et al., 2007]. The main difference is the presence of 2 additional arginines (Arg216 and Arg237) at the active site in substitution of a tyrosine (Tyr224) and a leucine (Leu215). This renders the active site of DASPO more positively charged, allowing an efficient binding and oxidation of the negatively charged D-aspartate.
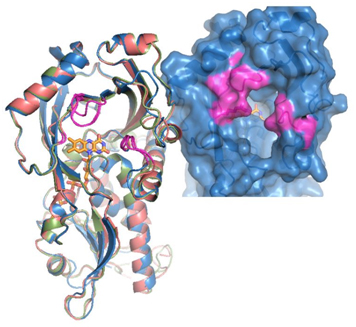
Fig. 3. 3D structure of mammalian DASPO.
Left: superimposition of the models of the 3D structures of DASPOs from rat (pink), mouse (blue), and human (green). Right: detail of the active site entrance of the human enzyme. The regions of the protein that slightly differ between the different mammalian DASPOs are highlighted in purple [Molla, unpublished].
DASPO as a novel drug target
Since D-Asp is involved in several crucial physiological and pathological processes of the central nervous system, it is not surprising that DASPO is considered a promising target for novel drug-based therapies for mental diseases [Errico et al., 2015b].
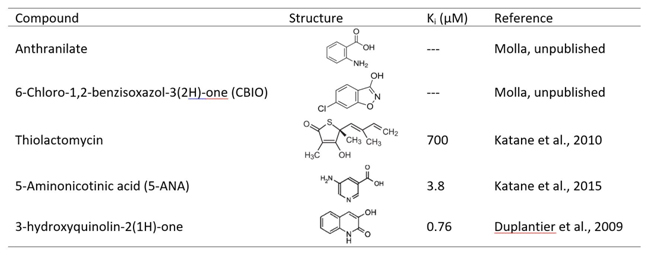
Inhibition of DASPO, in vivo, results in an increase in the concentration of free D-Asp, an effect that can be beneficial in those pathological conditions characterized by abnormally low concentrations of this amino acid [Errico et al., 2015b]. Activity of DASPO can be (competitively) inhibited by several classical amino acid oxidase inhibitors, such as tartrate, anthranilate, 6-chloro-1,2-benzisoxazol-3(2H)-one (CBIO), although with a very low potency (the IC50 is in the millimolar range). Recently, novel inhibitors with a Ki even in the nanomolar range have been identified: for example, thiolactomycin (a natural compound) or small aromatic compounds such as 5-aminonicotinic acid (5-ANA) or 3-hydroxyquinolin-2(1H)-one that have been discovered by a combined approach of in silico virtual screening and in vitro high-throughput screening [Katane et al., 2010b; Katane et al., 2015b; Duplantier et al., 2009) (Table 1).
Table 1. Selected inhibitors of DASPO.
Main properties (figures refers to human DASPO)
EC number: 1.4.3.1
Abbreviation: DASPO, DDO
Length (residues): 341
Mass (Da): 37535
Oligomerization state: Monomeric
Links
Brenda (http://www.brenda-enzymes.org/enzyme.php?ecno=1.4.3.1)
Wikipedia (https://en.wikipedia.org/wiki/D-aspartate_oxidase)
ExPASy (http://enzyme.expasy.org/EC/1.4.3.1)
UniProt (human) (http://www.uniprot.org/uniprot/Q99489)
References
Duplantier AJ, Becker SL, Bohanon MJ, Borzilleri KA, Chrunyk BA, Downs JT, Hu LY, El-Kattan A, James LC, Liu S, Lu J, Maklad N, Mansour MN, Mente S, Piotrowski MA, Sakya SM, Sheehan S, Steyn SJ, Strick CA, Williams VA, Zhang L., Discovery, SAR, and pharmacokinetics of a novel 3-hydroxyquinolin-2(1H)-one series of potent D-amino acid oxidase (DAAO) inhibitors. J. Med. Chem. 2009;52(11):3576-85.
Errico F, Mothet JP, Usiello A., D-Aspartate: An endogenous NMDA receptor agonist enriched in the developing brain with potential involvement in schizophrenia. J. Pharm. Biomed. Anal. 2015;116:7-17.
Errico F, D’Argenio V, Sforazzini F, Iasevoli F, Squillace M, Guerri G, Napolitano F, Angrisano T, Di Maio A, Keller S, Vitucci D, Galbusera A, Chiariotti L, Bertolino A, de Bartolomeis A, Salvatore F, Gozzi A, Usiello A., A role for D-aspartate oxidase in schizophrenia and in schizophrenia-related symptoms induced by phencyclidine in mice. Transl Psychiatry. 2015 17;5.
Katane M, Homma H., D-aspartate oxidase: the sole catabolic enzyme acting on free D-aspartate in mammals. Chem. Biodivers. 2017;7(6):1435-1449.
Katane M, Saitoh Y, Hanai T, Sekine M, Furuchi T, Koyama N, Nakagome I, Tomoda H, Hirono S, Homma H., Thiolactomycin inhibits D-aspartate oxidase: a novel approach to probing the active site environment. Biochimie. 2010;92(10):1371-1378.
Katane M, Kawata T, Nakayama K, Saitoh Y, Kaneko Y, Matsuda S, Saitoh Y, Miyamoto T, Sekine M, Homma H., Characterization of the enzymatic and structural properties of human D-aspartate oxidase and comparison with those of the rat and mouse enzymes. Biol Pharm Bull. 2015;38(2):298-305.
Katane M, Yamada S, Kawaguchi G, Chinen M, Matsumura M, Ando T, Doi I, Nakayama K, Kaneko Y, Matsuda S, Saitoh Y, Miyamoto T, Sekine M, Yamaotsu N, Hirono S, Homma H., Identification of novel D-aspartate oxidase inhibitors by in silico screening and their functional and structural characterization in vitro. J. Med. Chem. 2015;58(18):7328-7340.
Negri A, Tedeschi G, Ceciliani F, Ronchi S., 1999. Purification of beef kidney D-aspartate oxidase overexpressed in Escherichia coli and characterization of its redox potentials and oxidative activity towards agonists and antagonists of excitatory amino acid receptors. Biochimica et Biophysica Acta-Protein Structure and Molecular Enzymology 1431(1):212-222.
Pollegioni L, Piubelli L, Sacchi S, Pilone MS, Molla G., Physiological functions of D-amino acid oxidases: from yeast to humans. Cell Mol Life Sci. 2007;64(11):1373-94.
Umhau S, Pollegioni L, Molla G, Diederichs K, Welte W, Pilone MS, Ghisla S., The x-ray structure of D-amino acid oxidase at very high resolution identifies the chemical mechanism of flavin-dependent substrate dehydrogenation. Proc Natl Acad Sci U S A. 2000;97(23):12463-8.
Author
Gianluca Molla, Università degli studi dell’Insubria
